Simple Rigid Block Bonded-Block Modeling (BBM)
Problem Statement
Note
The project file for this example may be viewed/run in PFC.[1] The data file used is shown at the end of this example.
This example shows how to effectively use rigid blocks for simple Bonded-Block Modeling (BBM). The specimen created is shown below. This specimen is created using the rblock construct command. This command takes a geometry set and meshes it, creating a zero porosity packing of rigid blocks. The rigid blocks are then scaled so that they initially overlap. This is important for BBM modeling as, when rigid blocks are bonded, the computed contact location is only incrementally updated. In other words, the contact location does not
respond to changes in the overlap region. Instead, one can conceive of the contact location as the location
of an element of glue that is fixed to the surfaces of the rigid blocks until failure.
To dramatically increase computational performance, the rigid blocks are rounded with the rblock erode command. This command reduces the core shape and introduces rounding. This operation is important for rigid block modeling with zero porosity initial packings as the erosion removes many vertex-vertex and edge-edge contacts in the sense that they will be inactive. In fact, in this model with ~7700 rigid blocks there are ~13,000 face-face contacts between rigid blocks whereas there are more than 225,000 vertex-vertex and edge-edge contacts. These contacts contribute negligibly to test results but increase the computational time dramatically. In this model such contacts are inhibited with the contact inhibit command, meaning that they are skipped from all computations. If one were interested in very large strain where these contacts may be important, one could easily hook up a FISH function to the bond_break event and activate all or some subset of the contacts around the rigid blocks on each side of the contact.
The user must be aware that this procedure of requiring overlap for bonding results in the situation where if a contact model based on the Linear contact model is used the property lin_mode must be set to 1 so that normal forces are incrementally accumulated. Otherwise the initial overlap will result in initial contact forces that may lead to instability.

Figure 1: Initial specimen.
Simulation Results
A UCS and direct tension test are performed on the specimen. By specifying the initial velocities of the entire specimen and not just the platens, one can reduce any spurious oscillations in the stress-strain response. A FISH function is used to both compute the stress, strain and Young’s modulus and histories are kept of these quantities. The target material is a very brittle, hard rock with Young’s modulus 50 GPa, UCS strength ~ 110 MPa and tensile strength ~ 10 MPa. The Soft-Bond contact model is used without any softening for this simulation. With no softening this model is similar to the Linear Parallel Bond model in response with these notable exceptions: 1) the shear force is not reduced to 0 at shear failure but to the frictional sliding limit; 2) twisting resistance in 3D which is important for unbonded rigid blocks; 3) only one normal and shear stiffness is needed as opposed to separate parallel bond and linear stiffnesses.
The figures below demonstrate the UCS results run so that the residual strength is 80% of the peak. The desired peak value is achieved and the computed fragments show thorough failure of the specimen with both tensile and shear failures.
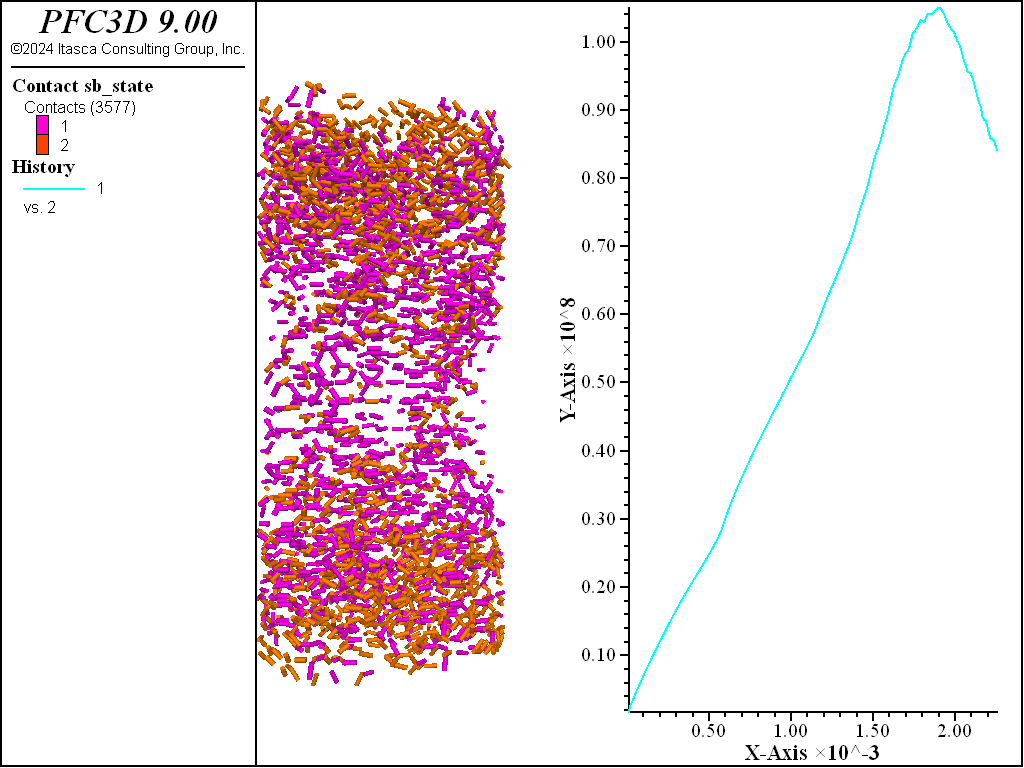
Figure 2: UCS results with failed bonds.
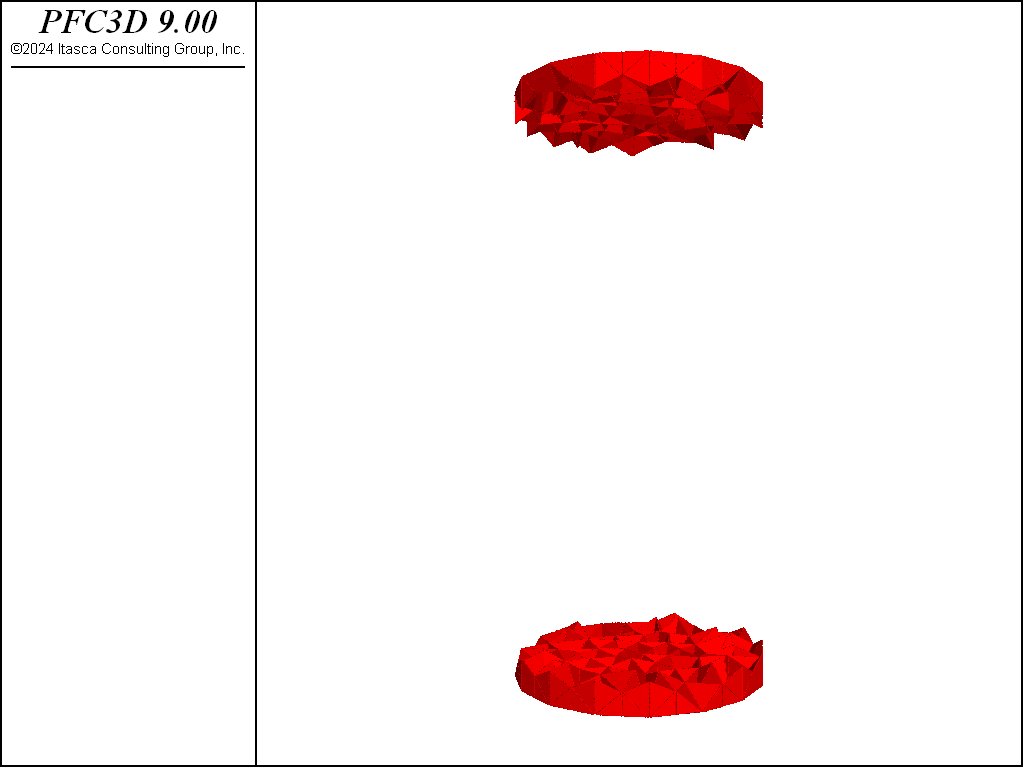
Figure 3: Fragments computed for the UCS test.
The figures below show the results of the direct tension test run so that the residual strength is 80% of the peak. Two distinct failure planes near the edges of the rigid blocks that are used as platens. The desired tensile strength is achieved.
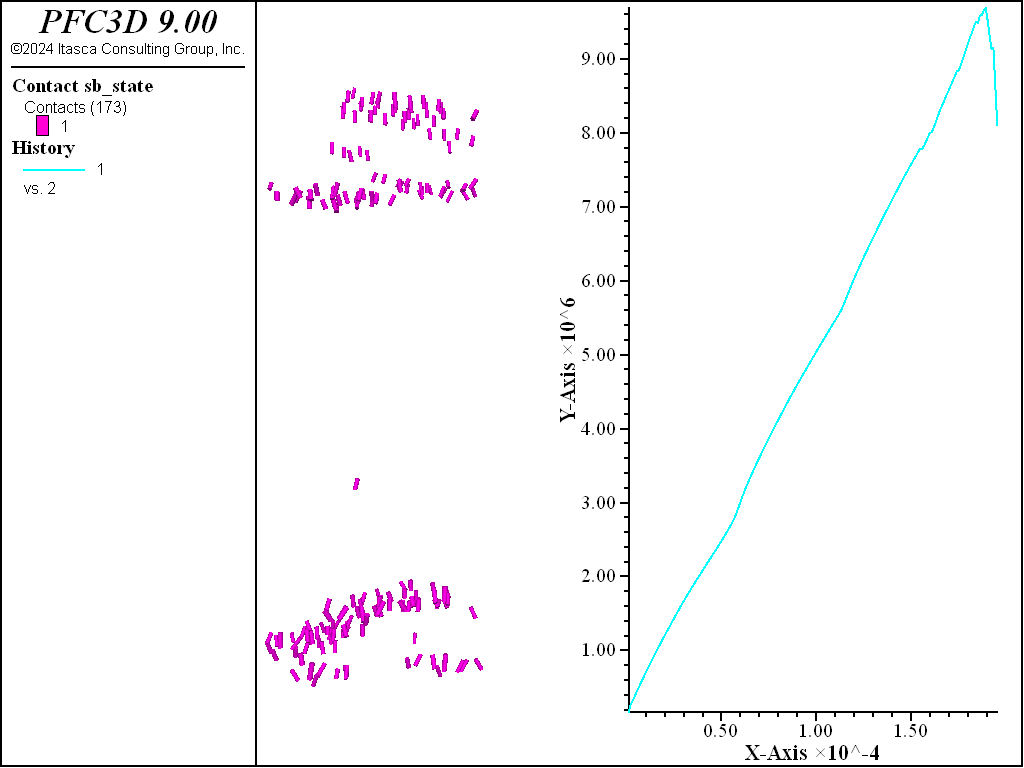
Figure 4: Direct tension results with failed bonds.
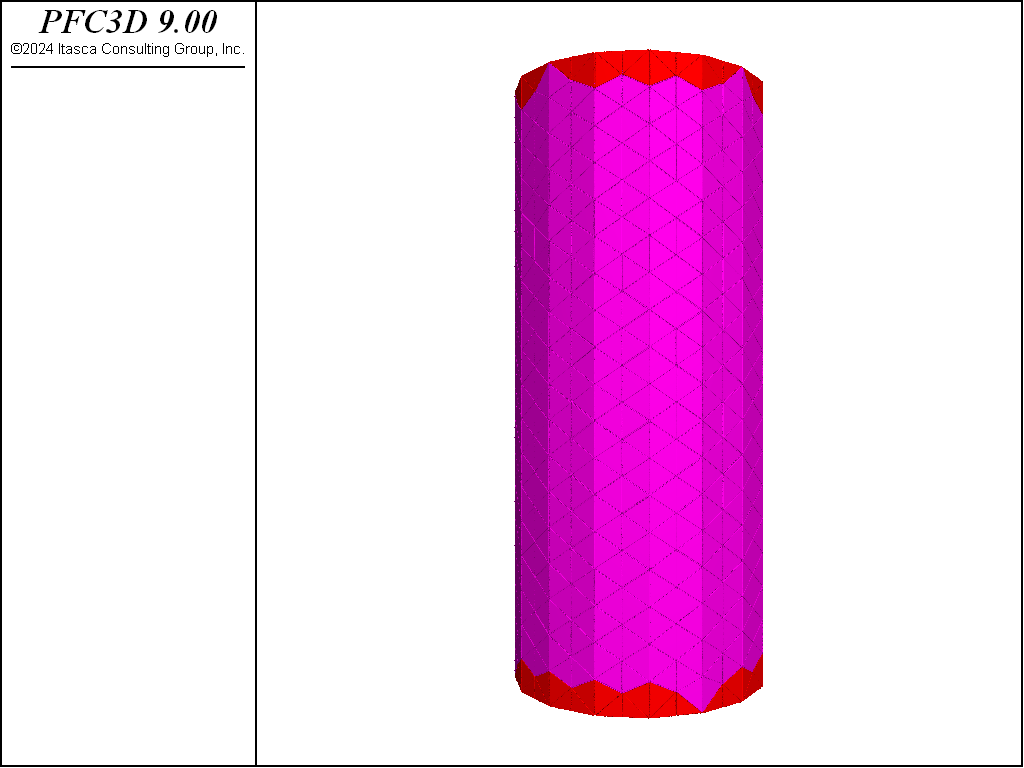
Figure 5: Fragments computed for the direct tension test.
Data Files
specimen.dat
; Clear the model state and create a domain in which all of the
; rigid blocks exist
model new
model large-strain on
model domain extent -10 10
; Create a cylindrical geometry set that is 2.5 times as tall as
; wide for the UCS and direct tension test
geometry set 'specimen'
[specRadius = 1.5]
[specHeight = 7.0]
geometry generate cylinder base 0 0 [-specHeight/2.0] radius [specRadius] ...
height [specHeight] axis 0 0 1 resolution 0.4
; Create the rigid blocks as tetrahedra. This command meshes the
; geometry set, creating rigid blocks.
rblock construct from-geometry 'specimen' maximum-edge 0.4
; Scale the rigid blocks so that they are originally overlapping. This is
; required for the contact location to be correct when bonding. When the bond
; method is called the contact location is only incrementally updated.
rblock scale relative 1.01
; Erode the rigid blocks, introducing rounding. This allows one to cull the
; numerous edge-edge and vertex-vertex contacts that do not contribute to the
; model response. This may fail at times so it is a good idea to use the
; skip-errors keyword
rblock erode relative 0.05 skip-errors
; Group the top and platens which are fixed chunks of rigid blocks.
rblock group 'top-platen' range position-z [specHeight/2.0 - 0.3] 10
rblock group 'bottom-platen' range position-z [0.3 - specHeight/2.0] -10
; Set the damping and density
rblock attribute density 2650 damp 0.7
; Fish symbol for the bond tensile strength
[tensileStrength = 4.7e6]
; Ratio cohesive to tensile strength
[cohesionFactor = 2.4]
; Target elastic modulus to put into the bonds
[emod = 7.25e10]
; Specify the softbond model properties.
contact cmat default model softbond method deformability emod [emod] ...
kratio 2.0 ...
property sb_ten [tensileStrength] ...
sb_coh [cohesionFactor*tensileStrength] ...
sb_fa 30.0 fric 0.5 sb_mode 1
; Create the contacts and bond them. Note that no softening is used in this
; example so the softbond contacts act essentially as parallel bond contacts
; except that there is only 1 set of stiffnesses and the shear stress is not
; nulled if shear failure occurs.
model clean all
contact method bond
; Make the platen contacts have the null contact model. This is to reduce
; computation and to ensure that they do not contribute to the stress
; computation. The matches keyword is used to specify that contacts fall within
; the range if both of the ends have the specified group.
contact model null range group 'top-platen' matches 2
contact model null range group 'bottom-platen' matches 2
; Inhibit the contacts that are not bonded. This is VERY important for
; efficient bonded ; block models. This means that the force-displacement law
; for edge-edge and vertex-vertex contacts is skipped along with updating the
; contact positions. You can compare the results and runtimes with and without
; the vertex-vertex and edge-edge contacts. If in small strain these contacts
; can actually be deleted with the contact delete command and contact
; detection can be turned off.
contact inhibit range contact gap 0 100
; Fix the velocity and spin of the top and bottom platens
rblock fix velocity spin range group 'top-platen'
rblock fix velocity spin range group 'bottom-platen'
; Make maps of the rigid blocks in the platens
[topMap = rblock.groupmap('top-platen')]
[botMap = rblock.groupmap('bottom-platen')]
; Define a FISH function to compute the stress and elastic modulus.
; Histories of these quantities will be kept for display.
[strain = 0.0]
[modulus = 0.0]
fish define stress
local stressTop = 0.0
local stressBot = 0.0
strain = 0.0
loop foreach b topMap
stressTop = stressTop + math.abs(rblock.force.contact(b,3))
strain = 2.*math.abs(rblock.disp(b,3))
endloop
loop foreach b botMap
stressBot = stressBot + math.abs(rblock.force.contact(b,3))
endloop
local st = (stressTop + stressBot)/(math.pi*specRadius^2)/2.0;
strain = strain/specHeight
if strain > 0
modulus = st / strain
endif
stress = st
end
fish history stress
fish history strain
fish history modulus
; Set up a FISH halt to end cycling when the stress is 80% of the maximum
[maxStress = 0.0]
[peakPercentage = 0.8]
fish define halt
halt = 0
maxStress = math.max(maxStress,stress)
if stress < peakPercentage * maxStress
halt = 1
endif
end
; Register for fragment computation
fragment register rblock-rblock
; Save the specimen
model save 'specimen'
tests.dat
;************************************************************
; UCS TEST
;************************************************************
; Restore the specimen and run the UCS
model restore 'specimen'
; Positive velocity means compression - this will be applied to the top
; and bottom platens
[nvel = 1.0]
; Apply the velocities to the rigid blocks so that
; the model isn't shocked by the initial velocity on the platens.
rblock attribute velocity-z 0.0 gradient 0 0 [-2.0 * nvel / specHeight]
; Set the platen velocities after assigning the velocities throughout
; the model.
rblock attribute velocity-z [nvel] range group 'bottom-platen'
rblock attribute velocity-z [-1*nvel] range group 'top-platen'
; Run the UCS test
model solve fish-halt halt cycles 1000000
; Compute fragments
fragment compute
; Save the UCS test
model save 'ucs'
;************************************************************
; DIRECT TENSION TEST
;************************************************************
; Restore the specimen and run the UCS
model restore 'specimen'
; Negative velocity means tension - this will be applied to the top
; and bottom platens
[nvel = -0.1]
; Apply the velocities to the rigid blocks so that
; the model isn't shocked by the initial velocity on the platens.
rblock attribute velocity-z 0.0 gradient 0 0 [-2.0 * nvel / specHeight]
; Set the platen velocities after assigning the velocities throughout
; the model.
rblock attribute velocity-z [nvel] range group 'bottom-platen'
rblock attribute velocity-z [-1*nvel] range group 'top-platen'
; Run the direct tension test
model solve fish-halt halt cycles 1000000
; Compute fragments
fragment compute
; Save the tension test
model save 'tension'
specimen.dat
; Clear the model state and create a domain in which all of the
; rigid blocks exist
model new
model large-strain on
model domain extent -10 10
; Create a cube of material for the UCS and direct tension test
geometry set 'specimen'
[specWidth = 7.0]
[specHeight = 7.0]
geometry generate box [-specWidth/2.0] [specWidth/2.0] ...
[-specHeight/2.0] [specHeight/2.0]
; Create the rigid blocks as tetrahedra. This command meshes the
; geometry set, creating rigid blocks.
rblock construct from-geometry 'specimen' maximum-edge 0.2
; Scale the rigid blocks so that they are originally overlapping. This is
; required for the contact location to be correct when bonding. When the bond
; method is called the contact location is only incrementally updated.
rblock scale relative 1.01
; Erode the rigid blocks, introducing rounding. This allows one to cull the
; numerous edge-edge and vertex-vertex contacts that do not contribute to the
; model response. This may fail at times so it is a good idea to use the
; skip-errors keyword
rblock erode relative 0.05 skip-errors
; Group the top and platens which are fixed chunks of rigid blocks.
rblock group 'top-platen' range position-y [specHeight/2.0 - 0.2] 10
rblock group 'bottom-platen' range position-y [0.2 - specHeight/2.0] -10
; Set the damping and density
rblock attribute density 2650 damp 0.7
; Fish symbol for the bond tensile strength
[tensileStrength = 2e7]
; Ratio cohesive to tensile strength
[cohesionFactor = 3.]
; Target elastic modulus to put into the bonds
[emod = 1.95e11]
; Specify the softbond model properties.
contact cmat default model softbond method deformability emod [emod] ...
kratio 2.0 ...
property sb_ten [tensileStrength] ...
sb_coh [cohesionFactor*tensileStrength] ...
sb_fa 30.0 fric 0.5 sb_mode 1
; Create the contacts and bond them. Note that no softening is used in this
; example so the softbond contacts act essentially as parallel bond contacts
; except that there is only 1 set of stiffnesses and the shear stress is not
; nulled if shear failure occurs.
model clean all
contact method bond
; Make the platen contacts have the null contact model. This is to reduce
; computation and to ensure that they do not contribute to the stress
; computation. The matches keyword is used to specify that contacts fall within
; the range if both of the ends have the specified group.
contact model null range group 'top-platen' matches 2
contact model null range group 'bottom-platen' matches 2
; Make the contacts with the platen rigid blocks have infinite strength
contact property sb_ten 1e10 sb_coh 1e10 range group 'top-platen' matches 1
contact property sb_ten 1e10 sb_coh 1e10 range group 'bottom-platen' matches 1
; Inhibit the contacts that are not bonded. This is VERY important for
; efficient bonded block models. This means that the force-displacement law
; for vertex-vertex contacts is skipped along with updating the contact
; positions. You can compare the results and runtimes with and without the
; vertex-vertex contacts. If in small strain these contacts can actually be
; deleted with the contact delete command and contact detection can be
; turned off.
contact inhibit range contact gap 0 100
; Fix the y-velocity of the top and bottom platens
rblock fix velocity-y spin range group 'top-platen'
rblock fix velocity-y spin range group 'bottom-platen'
; Make maps of the rigid blocks in the platens
[topMap = rblock.groupmap('top-platen')]
[botMap = rblock.groupmap('bottom-platen')]
; Define a FISH function to compute the stress and elastic modulus.
; Histories of these quantities will be kept for display.
[strain = 0.0]
[modulus = 0.0]
fish define stress
local stressTop = 0.0
local stressBot = 0.0
strain = 0.0
loop foreach b topMap
stressTop = stressTop + math.abs(rblock.force.contact(b,2))
strain = 2.*math.abs(rblock.disp(b,2))
endloop
loop foreach b botMap
stressBot = stressBot + math.abs(rblock.force.contact(b,2))
endloop
local st = (stressTop + stressBot)/2.0/specWidth
strain = strain/specHeight
if strain > 0
modulus = st / strain
endif
stress = st
end
fish history stress
fish history strain
fish history modulus
history interval 1
; Set up a FISH halt to end cycling when the stress is 80% of the maximum
[maxStress = 0.0]
[peakPercentage = 0.8]
fish define halt
halt = 0
maxStress = math.max(maxStress,stress)
if stress < peakPercentage * maxStress
halt = 1
endif
end
; Register for fragment computation
fragment register rblock-rblock
; Save the specimen
model save 'specimen'
tests.dat
;************************************************************
; UCS TEST
;************************************************************
; Restore the specimen and run the UCS
model restore 'specimen'
; Positive velocity means compression - this will be applied to the top
; and bottom platens
[nvel = 0.5]
; Apply the velocities to the rigid blocks so that
; the model isn't shocked by the initial velocity on the platens.
rblock attribute velocity-y 0.0 gradient 0 [-2.0 * nvel / specHeight]
; Set the platen velocities after assigning the velocities throughout
; the model.
rblock attribute velocity-y [nvel] range group 'bottom-platen'
rblock attribute velocity-y [-1*nvel] range group 'top-platen'
; Run the UCS test
model solve fish-halt halt cycles 1000000
; Compute fragments
fragment compute
; Save the UCS test
model save 'ucs'
;************************************************************
; DIRECT TENSION TEST
;************************************************************
; Restore the specimen and run the UCS
model restore 'specimen'
; Negative velocity means tension - this will be applied to the top
; and bottom platens
[nvel = -0.05]
; Apply the velocities to the rigid blocks so that
; the model isn't shocked by the initial velocity on the platens.
rblock attribute velocity-y 0.0 gradient 0 [-2.0 * nvel / specHeight]
; Set the platen velocities after assigning the velocities throughout
; the model.
rblock attribute velocity-y [nvel] range group 'bottom-platen'
rblock attribute velocity-y [-1*nvel] range group 'top-platen'
; Run the direct tension test
model solve fish-halt halt cycles 1000000
; Compute fragments
fragment compute
; Save the tension test
model save 'tension'
Endnotes
| Was this helpful? ... | Itasca Software © 2024, Itasca | Updated: Apr 02, 2024 |
