Spring Network Contact Model Capabilities
Problem Statement
Note
The project file for this example may be viewed/run in PFC.[1] The data files used are shown at the end of this example.
The Spring Network Model is an implementation of the Rigid Body Spring Network (RBSN) approach that is applicable to all PFC model components. This implementation provides some unique capabilities, including: 1) matching Young’s modulus based on lattice theory without calibration, 2) matching Poisson’s ratio, 3) reducing heterogeneity in elastic response, and 4) post-peak specification of tensile and shear strength as a function of displacement. This set of verification problems demonstrates some of these characteristics.
Matching Elastic Response
A tetrahedral packing composed of rigid blocks is created, trimmed to a cylindrical shape, and replicated. One specimen uses the Spring Network Model with the compute-stiffness method and the other uses the Soft-Bond Model model with the deformability method. The two specimen are squeezed quasi-statically and the stress/strain responses are recorded along with the Poisson’s ratios. Note that for soft-bonded material, the ratio of normal-to-shear stiffness is set to 2, which is the general rule of thumb for this contact model. The materials are bonded and failure is inhibited.
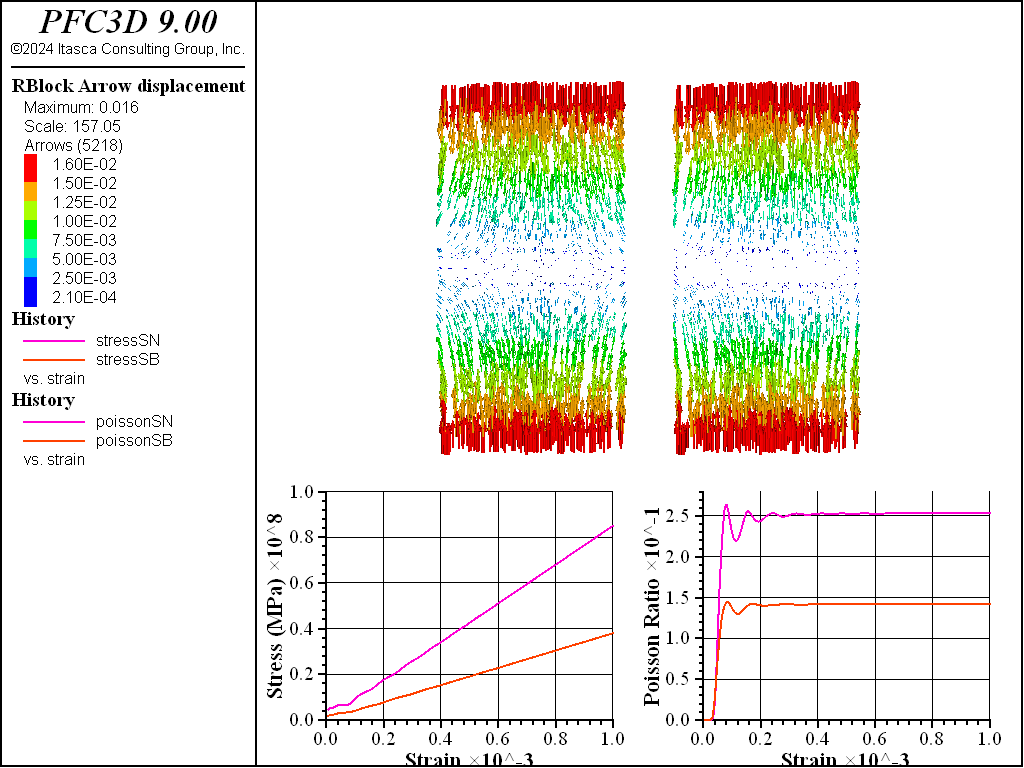
Figure 1: Stress-strain response, Poisson’s ratio, and displacement of the spring network and soft bond based specimen composed of tetrahedra.
As shown in the figure above, the stress-strain responses are both linear. But both the Young’s modulus and the Poisson’s ratio of the soft-bonded specimen are roughly half the spring network material values. In contrast, the Young’s modulus and Poisson’s ratio of the spring network material are both within ~ 5% of the target values. In tension, the behavior is the same. By trial and error, with this specific geometry, multiplication of the emod by 3.2 and setting the kratio to 3.5 in the deformability method matches the target material response, though these values depend on the specific geometry used in this example.
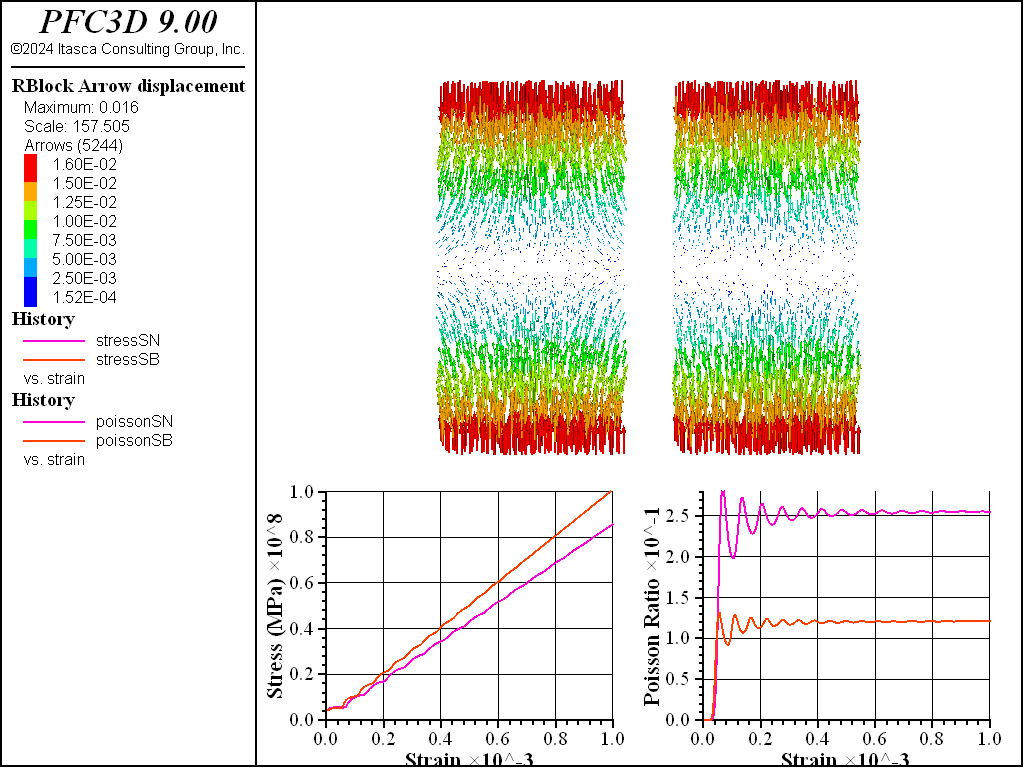
Figure 2: Stress-strain response, Poisson’s ratio, and displacement of the spring network and soft bond based specimen composed of Voronoi cells.
As the figure above shows, changing to a Voronoi packing results in a higher Young’s modulus for a soft-bonded material using the deformability method compared with the spring network material, though the Poisson’s ratio is still about half the target value, with a kratio of 2. Matching a target elastic response closely for a bonded specimen without calibration, regardless of packing, is a significant capability of the spring network model.
Importance of the Poisson Effect
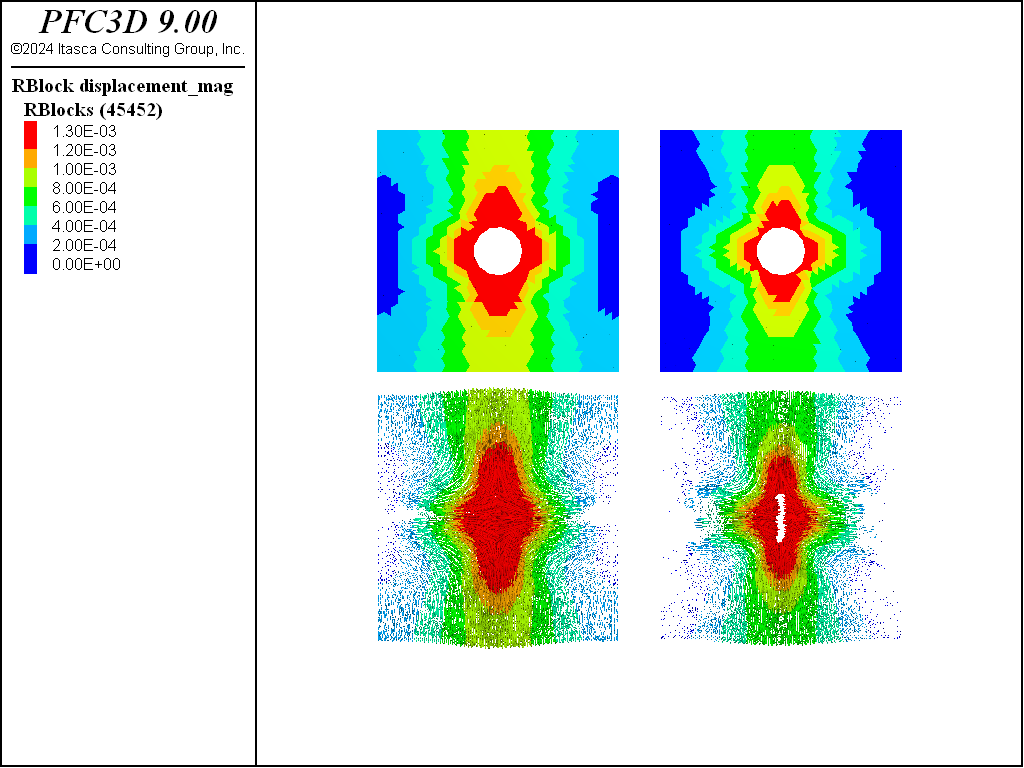
Matching the proper Poisson’s ratio can have important implications for stress re-distribution around openings in the presence of heterogeneous initial stresses. Note that for homogeneous stresses the impact of the Poisson effect is low. To demonstrate this effect, a circular hole is cut in an elastic material with and without the Poisson effect. The material is subjected to vertical tension and horizontal compression without failure. Stress boundaries are applied in the vertical direction while roller boundaries are applied in the horizontal directions.
The impact of the Poisson effect is apparent in the figure above, where vertical displacements are higher near the hole without Poisson ratio material. Thus the ability to match the macro Poisson effect directly may increase the fidelity of the model for such situations.
Tensile Softening
One way to boost the ratio of the unconfined compressive strength (UCS) to direct tensile strength in a bonded specimen is to introduce post-peak tensile softening. In the spring network model one can specify both a simple linear and a piecewise linear degradation of tensile force with continued elongation past the peak tensile strength. For linear softening one specifies the softening distance (sn_tendc property). For piecewise linear softening, table entries (sn_tentab property) can be specified as doublets of the ratio of the peak tensile force and distance. As with the soft bond model, if the bond undergoes compressive loading when in the softening regime, the stiffness reverts to the bond stiffness. If elongation resumes, the bond stiffness is used until the contact force reaches the previous softening curve, at which time softening resumes as specified.

Figure 4: Three contacts with: no tensile softening, linear tensile softening, and piecewise linear tensile softening.
The contacts are first loaded to failure and continue to elongate. Without tensile softening the tensile resistance to continued elongation drops instantaneously to 0. With tensile softening, tensile forces degrade as a function of elongation. The softening contacts are then exposed to compressive loading for a time, demonstrating that they follow the bonded stiffnesses when loading in compression. Subsequent tensile loading leads to continued softening once the stress-strain response intersects the previous softening curve. When the bond fails entirely, the Poisson correction is no longer computed; thus a new, frictional surface is created.
Slip Weakening Friction
The spring network model is capable of simulating both bonded materials and frictional surfaces. This example exercises frictional surfaces with varying frictional properties and constant normal stress. As with tensile softening, one may either specify a linear slip weakening distance (sn_cohdc property) or table entries for piecewise linear slip weakening (sn_cohtab property). In contrast to the tensile softening case, residual shear strength and healing may be specified. In all cases a friction angle of 15 degrees is used with a residual friction angle of 10 degrees (i.e., a residual friction coefficient ~0.176). A slide, hold, slide stress path is simulated for all contacts.
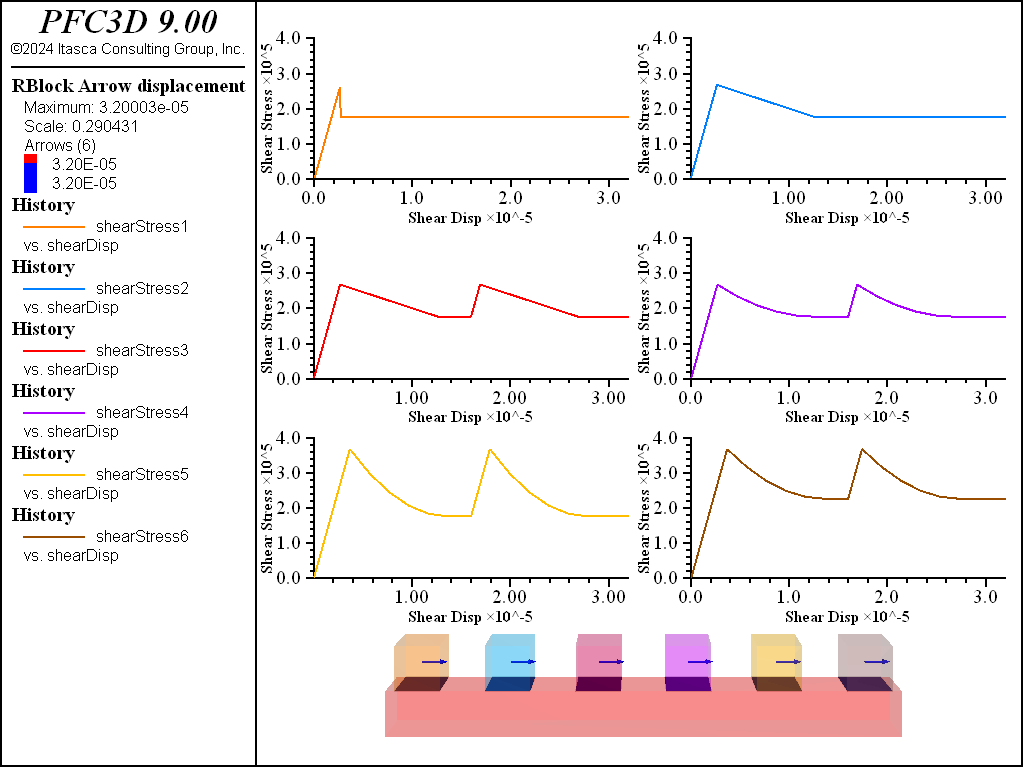
Figure 5: Six contacts with: 1) no slip weakening friction, 2) linear slip weakening friction, 3) linear slip weakening with healing, 4) piecewise linear slip weakening with healing, 5) piecewise linear slip weakening with healing and cohesion, 6) piecewise linear slip weakening with healing, cohesion, and residual cohesion.
Without weakening, bonding is required for the friction angle to contribute to the shear response. Without slip weakening, the shear stress drops instantaneously to 0. With slip weakening and without healing, the shear stress evolves to the sliding condition and sliding continues regardless of displacement history. Healing allows for the shear strength to reset once sliding ceases; this can allow for episodic slip to occur. It can be observed that piecewise linear slip weakening can also be specified, along with cohesive strength and residual cohesive strength.
Data Files
matchTet.dat (3D)
; Demonstrate matching the elastic constants for a tetrahedral
; packing
model new
model random 10000
; Use the facet area for the contact area, only make contacts with
; area greater than 1% of the total facet area of rblocks, and
; accmulate stresses for use with the springnetowrk model
rblock contact-resolution update-area true facet-total 0.01 ...
accumulate-stress true
model domain extent [-100] [100]
; Edge length for meshing
[maxEdge = 3.]
; Create a geometry set, mesh it, and remove delete the geometry set
geometry set 'blocks'
geometry generate box -30 -10 -10 10 -20 20
rblock construct from-geometry 'blocks' minimum-edge [maxEdge] ...
maximum-edge [maxEdge]
geometry delete
; Trim to a cylindrical container
fish define trim(h,r)
height = h
radius = r
command
geometry set 'container'
geometry generate cylinder resolution 0.5 ...
base -20 0 [-height/2.0] height [height] ...
radius [radius]
fracture import from-geometry 'container'
rblock cut fractures 'container' throughgoing ...
sliver-remove false group-facets 'outside' slot 'outside'
rblock delete range geometry-space 'container' count even
endcommand
; Compute the actual platen area
area = 0
local gs = geom.set.find('container')
loop foreach local p geom.poly.list(gs)
if math.dot(geom.poly.norm(p),vector(0,0,1)) > 0.98
area += geom.poly.area(p)
endif
endloop
command
geometry delete
fracture delete
endcommand
end
[trim(32,8)]
; Remove slivers
rblock delete aspect-ratio 10
rblock delete size absolute 1e-5
; Assign the group 'orig' to all rblocks
rblock group 'orig'
; FISH function to create copies of the previously created rigid blocks
; These rigid blocks are offset in the x direction by 20 units and a group
; is assigned
fish define replicate
xoffset = 20
loop foreach local rb rblock.group.list('orig')
local rb2 = rblock.copy(rb)
rblock.pos(rb2)->x += xoffset
rblock.group(rb2) = 'replicated'
endloop
end
[replicate]
; Assign groups for histories. SN stands for springnetwork while SB
; Stands for softbond
rblock group 'bottomSN' slot 'gauge' range ...
position-z -16 by rblock-vertex group 'orig'
rblock group 'topSN' slot 'gauge' range ...
position-z 16 by rblock-vertex group 'orig'
rblock group 'bottomSB' slot 'gauge' range ...
position-z -16 by rblock-vertex group 'replicated'
rblock group 'topSB' slot 'gauge' range ...
position-z 16 by rblock-vertex group 'replicated'
rblock group 'poissonSN' slot 'gauge' range ...
group 'outside' slot 'outside' by rblock-facet ...
position-z -1 1 by rblock-vertex group 'orig'
rblock group 'poissonSB' slot 'gauge' range ...
group 'outside' slot 'outside' by rblock-facet ...
position-z -1 1 by rblock-vertex group 'replicated'
; Fix the velocity and spin of the top and bottom blocks
rblock fix velocity spin range ...
group 'bottomSN' slot 'gauge' ...
group 'topSN' slot 'gauge' union
rblock fix velocity spin range ...
group 'bottomSB' slot 'gauge' ...
group 'topSB' slot 'gauge' union
; Young's modulus, Poisson ratio and density
[E = 8e10]
[pois = 0.25]
[den = 2670.]
rblock attribute density [den] damp 0.7
; Dilate but keep inertial attributes unchanged for contacts detection.
rblock dilate expand 0.0001 keep-inertial
; Default CMAT is springnetwork
contact cmat default model springnetwork ...
method compute-stiffness emod [E] poisson [pois] ...
property sn_ten 1e100 sn_coh 1e100
; Create contacts
model clean all
; For the replicated rblocks use the softbond model with deformability
; method and kratio 2; give them infinite strength as well.
contact model softbond range group 'replicated' matches 2
contact method deformability emod [E] kratio 2.0
contact property sb_coh 1e100 sb_ten 1e100
; Bond all contacts, set to large strain, and timestep scaling
contact method bond
model large-strain on
model mechanical timestep scale
; Get lists of rblocks for histories.
[topSN = rblock.group.list('topSN','gauge')]
[botSN = rblock.group.list('bottomSN','gauge')]
[topSB = rblock.group.list('topSB','gauge')]
[botSB = rblock.group.list('bottomSB','gauge')]
[poisSN = rblock.group.list('poissonSN','gauge')]
[poisSB = rblock.group.list('poissonSB','gauge')]
; Centers of specimen for Poisson ratio computation
[cenSNx = -20]
[cenSBx = 0]
; FISH histories
fish define strain
theStrain = math.abs(rblock.disp(topSN(1))->z)*2.0/height
strain = theStrain
stressSN = 0
loop foreach local rb topSN
stressSN += rblock.force.unbal(rb)->z
endloop
loop foreach rb botSN
stressSN -= rblock.force.unbal(rb)->z
endloop
stressSN = math.abs(stressSN)/2.0/area
stressSB = 0
loop foreach rb topSB
stressSB += rblock.force.unbal(rb)->z
endloop
loop foreach rb botSB
stressSB -= rblock.force.unbal(rb)->z
endloop
stressSB = math.abs(stressSB)/2.0/area
poissonSN = 0
poissonSB = 0
if theStrain # 0
local cd = 0
loop foreach rb poisSN
local vcomp = rblock.pos(rb)
local op = vector(cenSNx,0,vcomp->z)
cd = cd + math.dot(math.unit(vcomp-op),rblock.disp(rb))
endloop
cd /= list.size(poisSN)
cd = cd/radius
poissonSN = math.abs(cd/theStrain)
cd = 0
loop foreach rb poisSB
vcomp = rblock.pos(rb)
op = vector(cenSBx,0,vcomp->z)
cd = cd + math.dot(math.unit(vcomp-op),rblock.disp(rb))
endloop
cd /= list.size(poisSB)
cd = cd/radius
poissonSB = math.abs(cd/theStrain)
endif
end
fish history name 'strain' strain
fish history name 'stressSN' stressSN
fish history name 'stressSB' stressSB
fish history name 'poissonSN' poissonSN
fish history name 'poissonSB' poissonSB
model save 'beforeSqueezeTet'
model restore 'beforeSqueezeTet'
; Set the velocity of throughout the material to reduce oscillations.
rblock attribute velocity-z 0 gradient 0 0 [-1e-5/(height/2.0)]
; Set the platen velocities directly
rblock attribute velocity-z 1e-5 range ...
group 'bottomSN' slot 'gauge' ...
group 'bottomSB' slot 'gauge' union
rblock attribute velocity-z -1e-5 range ...
group 'topSN' slot 'gauge' ...
group 'topSB' slot 'gauge' union
model cycle 1600
model save 'afterSqueezeTet'
matchVor.dat (3D)
; Demonstrate matching the elastic constants for a Voronoi
; packing
model new
model random 10000
; Use the facet area for the contact area, only make contacts with
; area greater than 1% of the total facet area of rblocks, and
; accmulate stresses for use with the springnetowrk model
rblock contact-resolution update-area true facet-total 0.01 ...
accumulate-stress true
model domain extent [-100] [100]
; Create balls for Voronoi tessellation
ball generate number 100000 radius 0.5 0.6 box -30 -10 -10 10 -20 20
; Construct rblocks
rblock construct from-balls polydisperse true
; Delete balls
ball delete
; Trim to a cylindrical container
fish define trim(h,r)
height = h
radius = r
command
geometry set 'container'
geometry generate cylinder resolution 0.5 ...
base -20 0 [-height/2.0] height [height] ...
radius [radius]
fracture import from-geometry 'container'
rblock cut fractures 'container' throughgoing ...
sliver-remove false group-facets 'outside' slot 'outside'
rblock delete range geometry-space 'container' count even
endcommand
; Compute the actual platen area
area = 0
local gs = geom.set.find('container')
loop foreach local p geom.poly.list(gs)
if math.dot(geom.poly.norm(p),vector(0,0,1)) > 0.98
area += geom.poly.area(p)
endif
endloop
command
geometry delete
fracture delete
endcommand
end
[trim(32,8)]
; Remove slivers
rblock delete aspect-ratio 10
rblock delete size absolute 1e-5
; Assign the group 'orig' to all rblocks
rblock group 'orig'
; FISH function to create copies of the previously created rigid blocks
; These rigid blocks are offset in the x direction by 20 units and a group
; is assigned
fish define replicate
xoffset = 20
loop foreach local rb rblock.group.list('orig')
local rb2 = rblock.copy(rb)
rblock.pos(rb2)->x += xoffset
rblock.group(rb2) = 'replicated'
endloop
end
[replicate]
; Assign groups for histories. SN stands for springnetwork while SB
; Stands for softbond
rblock group 'bottomSN' slot 'gauge' range ...
position-z -16 by rblock-vertex group 'orig'
rblock group 'topSN' slot 'gauge' range ...
position-z 16 by rblock-vertex group 'orig'
rblock group 'bottomSB' slot 'gauge' range ...
position-z -16 by rblock-vertex group 'replicated'
rblock group 'topSB' slot 'gauge' range ...
position-z 16 by rblock-vertex group 'replicated'
rblock group 'poissonSN' slot 'gauge' range ...
group 'outside' slot 'outside' by rblock-facet ...
position-z -1 1 by rblock-vertex group 'orig'
rblock group 'poissonSB' slot 'gauge' range ...
group 'outside' slot 'outside' by rblock-facet ...
position-z -1 1 by rblock-vertex group 'replicated'
; Fix the velocity and spin of the top and bottom blocks
rblock fix velocity spin range ...
group 'bottomSN' slot 'gauge' ...
group 'topSN' slot 'gauge' union
rblock fix velocity spin range ...
group 'bottomSB' slot 'gauge' ...
group 'topSB' slot 'gauge' union
; Young's modulus, Poisson ratio and density
[E = 8e10]
[pois = 0.25]
[den = 2670.]
rblock attribute density [den] damp 0.7
; Dilate but keep inertial attributes unchanged for contacts detection.
rblock dilate expand 0.0001 keep-inertial
; Default CMAT is springnetwork
contact cmat default model springnetwork ...
method compute-stiffness emod [E] poisson [pois] ...
property sn_ten 1e100 sn_coh 1e100
; Create contacts
model clean all
; For the replicated rblocks use the softbond model with deformability
; method and kratio 2; give them infinite strength as well.
contact model softbond range group 'replicated' matches 2
contact method deformability emod [E] kratio 2.0
contact property sb_coh 1e100 sb_ten 1e100
; Bond all contacts, set to large strain, and timestep scaling
contact method bond
model large-strain on
model mechanical timestep scale
; Get lists of rblocks for histories.
[topSN = rblock.group.list('topSN','gauge')]
[botSN = rblock.group.list('bottomSN','gauge')]
[topSB = rblock.group.list('topSB','gauge')]
[botSB = rblock.group.list('bottomSB','gauge')]
[poisSN = rblock.group.list('poissonSN','gauge')]
[poisSB = rblock.group.list('poissonSB','gauge')]
; Centers of specimen for Poisson ratio computation
[cenSNx = -20]
[cenSBx = 0]
; FISH histories
fish define strain
theStrain = math.abs(rblock.disp(topSN(1))->z)*2.0/height
strain = theStrain
stressSN = 0
loop foreach local rb topSN
stressSN += rblock.force.unbal(rb)->z
endloop
loop foreach rb botSN
stressSN -= rblock.force.unbal(rb)->z
endloop
stressSN = math.abs(stressSN)/2.0/area
stressSB = 0
loop foreach rb topSB
stressSB += rblock.force.unbal(rb)->z
endloop
loop foreach rb botSB
stressSB -= rblock.force.unbal(rb)->z
endloop
stressSB = math.abs(stressSB)/2.0/area
poissonSN = 0
poissonSB = 0
if theStrain # 0
local cd = 0
loop foreach rb poisSN
local vcomp = rblock.pos(rb)
local op = vector(cenSNx,0,vcomp->z)
cd = cd + math.dot(math.unit(vcomp-op),rblock.disp(rb))
endloop
cd /= list.size(poisSN)
cd = cd/radius
poissonSN = math.abs(cd/theStrain)
cd = 0
loop foreach rb poisSB
vcomp = rblock.pos(rb)
op = vector(cenSBx,0,vcomp->z)
cd = cd + math.dot(math.unit(vcomp-op),rblock.disp(rb))
endloop
cd /= list.size(poisSB)
cd = cd/radius
poissonSB = math.abs(cd/theStrain)
endif
end
fish history name 'strain' strain
fish history name 'stressSN' stressSN
fish history name 'stressSB' stressSB
fish history name 'poissonSN' poissonSN
fish history name 'poissonSB' poissonSB
model save 'beforeSqueezeVor'
model restore 'beforeSqueezeVor'
; Set the velocity of throughout the material to reduce oscillations.
rblock attribute velocity-z 0 gradient 0 0 [-1e-5/(height/2.0)]
; Set the platen velocities directly
rblock attribute velocity-z 1e-5 range ...
group 'bottomSN' slot 'gauge' ...
group 'bottomSB' slot 'gauge' union
rblock attribute velocity-z -1e-5 range ...
group 'topSN' slot 'gauge' ...
group 'topSB' slot 'gauge' union
model cycle 1600
model save 'afterSqueezeVor'
simpleHole.dat (3D)
; Demonstrate Poisson effect for a circular hole in an elastic
; plate.
model new
model random 10000
; Use the facet area for the contact area, only make contacts with
; area greater than 1% of the total facet area of rblocks, and
; accmulate stresses for use with the springnetowrk model
rblock contact-resolution update-area true facet-total 0.01 ...
accumulate-stress true
; Specimen dimensions
[radius = 10.]
[length = 100.]
[width = 10.]
model domain extent [-4*length] [4*length]
; Offset of specimen in the x-direction.
[xoffset = -length/1.5]
; Edge length for constructing rblocks.
[maxEdge = width/3.]
; Construct rblocks from a geometry set.
geometry set 'blocks'
geometry generate box [-length/2.0 + xoffset] [length/2.0 + xoffset] ...
[-width/2.0] [width/2.0] [-length/2.0] [length/2.0]
rblock construct from-geometry 'blocks' ...
minimum-edge [maxEdge/2.0] maximum-edge [maxEdge]
geometry delete
; Create the hole and cut the rblocks, leaving the slivers.
geometry set 'hole'
geometry generate cylinder resolution 0.5 ...
base [xoffset] [-width] 0 axis 0 1 0 height [2*width] ...
radius [radius]
fracture import from-geometry 'hole'
rblock cut fractures 'hole' throughgoing ...
sliver-remove false
; Assign a group to the rblocks in the hole
rblock group 'hole' slot 'hole' range geometry-space 'hole' count odd
geometry delete
fracture delete
; Remove slivers
rblock delete aspect-ratio 10
rblock delete size absolute 1e-5
; Group all rblocks as 'orig' for replciation.
rblock group 'orig'
; Assign boundary groups to facets
rblock facet group 'yb' slot 'boundary' range ...
position-y [width/2.0]
rblock facet group 'yb' slot 'boundary' range ...
position-y [-width/2.0]
rblock facet group 'zb' slot 'boundary' range ...
position-z [length/2.0]
rblock facet group 'zb' slot 'boundary' range ...
position-z [-length/2.0]
rblock facet group 'xb' slot 'boundary' range ...
position-x [length/2.0 + xoffset]
rblock facet group 'xb' slot 'boundary' range ...
position-x [-length/2.0 + xoffset]
; Replicate the original rblocks and move them in the
; x-direction. Assign a different group for identification.
fish define replicate
loop foreach local rb rblock.group.list('orig')
local rb2 = rblock.copy(rb)
rblock.pos(rb2)->x += length/2.0 - xoffset
rblock.group(rb2) = 'replicated'
endloop
end
[replicate]
model save 'holeModel'
model restore 'holeModel'
; Stress tensor for confinement (negative is compression).
[confine = tensor(-10e6,-10e6,10e6,0.0,0.0,0.0)]
; Apply the stress tensor to the boundary blocks in the z-direction.
rblock facet apply stress [confine->xx] [confine->yy] [confine->zz] ...
[confine->xy] [confine->xz] [confine->yz] ...
range group 'zb' slot 'boundary'
; Make the x- and y-direction boundaries roller boundaries.
rblock fix velocity-x spin range group 'xb' slot 'boundary' ...
by rblock-facet
rblock fix velocity-y spin range group 'yb' slot 'boundary' ...
by rblock-facet
; Material properties
[E = 8e10]
[pois = 0.25]
[den = 2670.]
rblock attribute density [den] damp 0.7
; Dilate but keep inertial attributes unchanged for contacts detection.
rblock dilate expand 0.001 keep-inertial
; Default CMAT.
contact cmat default model springnetwork ...
method compute-stiffness emod [E] poisson [pois] ...
property sn_ten 1e100 sn_coh 1e100
model clean all
; Bond all contacts.
contact method bond
; Set the original rblocks to have 0 Poisson ratio.
contact property sn_pois 0 range group 'orig' matches 2
; Install stresses in the rblocks.
rblock tractions constant [confine->xx] [confine->yy] [confine->zz] ...
[confine->xy] [confine->xz] [confine->yz]
; Timestep scaling and large strain enabled.
model mechanical timestep scale
model large-strain on
; Solve to average ratio 1e-5
model solve
; Remove the hole material and reset the displacements.
rblock delete range group 'hole' slot 'hole'
rblock attribute displacement 0 0 0
; Solve again to average ratio 1e-5
model cycle 1
model solve
model save 'holeFinal'
tension.dat (3D)
; Demonstrate tensile response with various model parameters.
model new
model domain extent -10 10
; Use the facet area for the contact area, only make contacts with
; area greater than 1% of the total facet area of rblocks, and
; accmulate stresses for use with the springnetowrk model
rblock contact-resolution update-area true facet-total 0.01 ...
accumulate-stress true
; Create a single block for the base.
rblock create box -1 1.5 -0.25 0.25 -0.5 0 group 'base'
; Create individual rblocks to test different properties.
rblock create box -1 -0.5 -0.25 0.25 -1e-5 0.5 group 'slider1'
rblock create box 0 0.5 -0.25 0.25 -1e-5 0.5 group 'slider2'
rblock create box 1 1.5 -0.25 0.25 -1e-5 0.5 group 'slider3'
; Assign density and damping.
rblock attribute density 2000 damp 0.7
; Fix the velocity and all rblocks.
rblock fix velocity spin
; All contacts are springnetwork with stiffness/area 1e11 in normal and shear.
contact cmat default model springnetwork ...
method assign-stiffness kn 1e11 kratio 1 ...
property sn_coh 1e100 sn_ten 1e6
; Put in a compressive stress.
rblock tractions constant 0 0 -1e-6 0 0 0
; Run these in large strain.
model large-strain on
; Use FISH histories to track the shear stress and shear displacements
; of each contact.
; Since all rblocks have fixed velocities then the displacements are the same.
fish define normStress1
normStress1 = contact.prop(contact.find('rblock-rblock',1),'sn_sigma')
normStress2 = contact.prop(contact.find('rblock-rblock',2),'sn_sigma')
normStress3 = contact.prop(contact.find('rblock-rblock',3),'sn_sigma')
normDisp = rblock.disp(rblock.find(2))->z
end
; Create the histories
fish history name 'normStress1' normStress1
fish history name 'normStress2' normStress2
fish history name 'normStress3' normStress3
fish history name 'normDisp' normDisp
; Record the histories every 10 cycles.
history interval 10
; Set the x-velocity of the rblocks.
rblock attribute velocity-z 1e-4 range group 'base' not
; Create all contacts and bond them.
model clean all
contact method bond
; Second block - add linear softening.
contact property sn_tendc 1e-5 range group 'slider2'
; Third block - non-linear softening.
contact property sn_tendc 1e-5 ...
sn_tentab [vector(0.8^2,1e-5*(1-0.8))] sn_tentab [vector(0.6^2,1e-5*(1-0.6))] ...
sn_tentab [vector(0.4^2,1e-5*(1-0.4))] sn_tentab [vector(0.2^2,1e-5*(1-0.2))] ...
range group 'slider3'
; Since all rblocks are fixed need to set a timestep.
model mechanical timestep fix 1.5e-4
; Elongate.
model cycle 1000
; Compress.
rblock attribute velocity-z -1e-4 range group 'base' not
model cycle 200
; Elongate again.
rblock attribute velocity-z 1e-4 range group 'base' not
model cycle 2500
model save 'tensionFinal'
shear.dat (3D)
; Demonstrate shear response with various model parameters.
model new
model domain extent -10 10
; Use the facet area for the contact area, only make contacts with
; area greater than 1% of the total facet area of rblocks, and
; accmulate stresses for use with the springnetowrk model.
rblock contact-resolution update-area true facet-total 0.01 ...
accumulate-stress true
; Create a single block for the base.
rblock create box -3.1 2.6 -0.25 0.25 -0.5 0 group 'base'
; Create individual sliders to test different properties.
rblock create box -3 -2.5 -0.25 0.25 -1e-5 0.5 group 'slider1'
rblock create box -2 -1.5 -0.25 0.25 -1e-5 0.5 group 'slider2'
rblock create box -1 -0.5 -0.25 0.25 -1e-5 0.5 group 'slider3'
rblock create box 0 0.5 -0.25 0.25 -1e-5 0.5 group 'slider4'
rblock create box 1 1.5 -0.25 0.25 -1e-5 0.5 group 'slider5'
rblock create box 2 2.5 -0.25 0.25 -1e-5 0.5 group 'slider6'
; Assign density and damping.
rblock attribute density 2000 damp 0.7
; Fix the velocity and spin of the base, and the x-velocity
; and spin of the sliders.
rblock fix velocity spin range group 'base'
rblock fix velocity-x spin range group 'base' not
; Apply a normal stress to the top face of the sliders.
rblock facet apply stress-normal -1.e6 range ...
position-z 0.5 tolerance 1e-3
; All contacts are springnetwork with stiffness/area 1e11 in normal
; and shear.
contact cmat default model springnetwork ...
method assign-stiffness kn 1e11 kratio 1
; Put in a compressive stress.
rblock tractions constant 0 0 -1e-6 0 0 0
; Run these in large strain.
model large-strain on
; Solve to an average ratio of 1e-5.
model solve
; Reset all of the displacements in the springnetwork contacts and ...
; rblock displacements.
contact method reset-disp
rblock attribute displacement 0 0 0
; Fix the vertical velocity
rblock attribute velocity 0 0 0
rblock fix velocity-z
; Use FISH histories to track the shear stress and shear displacement
; of each contact.
; Since all rblocks have fixed velocities then the displacements are the same.
fish define shearStress1
shearStress1 = contact.prop(contact.find('rblock-rblock',1),'sn_tau')
shearStress2 = contact.prop(contact.find('rblock-rblock',2),'sn_tau')
shearStress3 = contact.prop(contact.find('rblock-rblock',3),'sn_tau')
shearStress4 = contact.prop(contact.find('rblock-rblock',4),'sn_tau')
shearStress5 = contact.prop(contact.find('rblock-rblock',5),'sn_tau')
shearStress6 = contact.prop(contact.find('rblock-rblock',6),'sn_tau')
shearDisp = contact.prop(contact.find('rblock-rblock',1),'sn_sdisp')->y
end
; Create the histories
fish history name 'shearStress1' shearStress1
fish history name 'shearStress2' shearStress2
fish history name 'shearStress3' shearStress3
fish history name 'shearStress4' shearStress4
fish history name 'shearStress5' shearStress5
fish history name 'shearStress6' shearStress6
fish history name 'shearDisp' shearDisp
; Record the histories every 10 cycles.
history interval 10
; Set the x-velocity of the sliders.
rblock attribute velocity-x 1e-4 range group 'base' not
; Bond all contacts.
contact method bond
; All contacts have the same sn_fa and fric, here in degrees.
[fricAngle = 15.]
[fricRes = 10.]
contact property sn_fa [fricAngle] fric [math.tan(fricRes/180*math.pi)]
; Second block - add sn_cohdc.
contact property sn_cohdc 1e-5 range group 'slider2'
; Third block - add sn_cohdc, healing.
contact property sn_cohdc 1e-5 sn_heal 1 range group 'slider3'
; Fourth block - non-linear, healing.
contact property sn_cohdc 1e-5 sn_heal 1 ...
sn_cohtab [vector(0.8^2,1e-5*(1-0.8))] sn_cohtab [vector(0.6^2,1e-5*(1-0.6))] ...
sn_cohtab [vector(0.4^2,1e-5*(1-0.4))] sn_cohtab [vector(0.2^2,1e-5*(1-0.2))] ...
range group 'slider4'
; Fifth block - non-linear, healing, cohesion.
contact property sn_cohdc 1e-5 sn_heal 1 ...
sn_cohtab [vector(0.8^2,1e-5*(1-0.8))] sn_cohtab [vector(0.6^2,1e-5*(1-0.6))] ...
sn_cohtab [vector(0.4^2,1e-5*(1-0.4))] sn_cohtab [vector(0.2^2,1e-5*(1-0.2))] ...
sn_coh 1e5 range group 'slider5'
; Sixth block - non-linear, healing, cohesion, residual cohesion.
contact property sn_cohdc 1e-5 sn_heal 1 ...
sn_cohtab [vector(0.8^2,1e-5*(1-0.8))] sn_cohtab [vector(0.6^2,1e-5*(1-0.6))] ...
sn_cohtab [vector(0.4^2,1e-5*(1-0.4))] sn_cohtab [vector(0.2^2,1e-5*(1-0.2))] ...
sn_coh 1e5 sn_cohres 5e4 range group 'slider6'
; Displace in shear.
model cycle 2000
; Stop sliding by setting the velocity to 0 and cycling.
rblock attribute velocity-x 0 range group 'base' not
model cycle 200
; Displace in shear again.
rblock attribute velocity-x 1e-4 range group 'base' not
model cycle 2000
model save 'shearFinal'
Endnote
| Was this helpful? ... | Itasca Software © 2024, Itasca | Updated: Nov 12, 2025 |